15 May 2021 - {{hitsCtrl.values.hits}}

COVID-19 patients receive oxygen outside an overwhelmed hospital in Kathmandu, Nepal.Credit: Navesh Chitrakar/Reuters
From Sri Lanka to Nepal, scientists with limited resources are working feverishly to discover which variants are driving outbreaks
Especially worrying is B.1.617, which was first identified in the Indian state of Maharashtra, and which the World Health Organization has said is probably highly transmissible
Malavige from May is doubling the average number of samples sequenced each month to nearly 200
 In early April, Neelika Malavige shut down her laboratory in Sri Lanka for ten days to celebrate Sinhalese New Year. Malavige, an immunologist at the University of Sri Jayewardenepura in Colombo, runs the only lab in the nation that sequences SARS-CoV-2 genomes. But case counts had dipped, hotspots had started to receive vaccines, and her team needed the break.
In early April, Neelika Malavige shut down her laboratory in Sri Lanka for ten days to celebrate Sinhalese New Year. Malavige, an immunologist at the University of Sri Jayewardenepura in Colombo, runs the only lab in the nation that sequences SARS-CoV-2 genomes. But case counts had dipped, hotspots had started to receive vaccines, and her team needed the break.
When the researchers returned to work, however, what they found was dizzying. Of 78 samples that they sequenced from people with COVID-19 in late April, 66 contained the highly transmissible B.1.1.7 variant. Since April 17, the number of new COVID-19 cases in Sri Lanka — which hit 2,672 on May 9 — has been breaking records almost every day. Hospitals are filling up and Malavige is bracing for a looming wave of deaths. “The situation in Sri Lanka is looking very grim,” she says.
As India struggles under the weight of a massive surge of COVID-19, many of its neighbours in south and southeast Asia are experiencing some of their largest outbreaks yet (see ‘Asia’s growing outbreaks’). There are even surges in Bhutan, where 62% of the population has received at least one dose of a vaccine, and Laos, which has previously had very few cases — although these outbreaks are small compared with those in nearby nations.
Spillover from India?
|
It’s important to ramp up our sequencing to make sure we detect these variants in the community as soon as possible |
Researchers region-wide are embarking on a remarkable effort to quickly scale up their very limited sequencing capacities to identify whether India’s epidemic — fuelled by variants such as B.1.617 — is spilling over into their communities, or if their outbreaks have origins elsewhere. In some cases, they are working around the clock and with insufficient resources. Specifically, they want to know whether worrying variants are circulating, so that they can assess the risks.
“It’s important to ramp up our sequencing to make sure we detect these variants in the community as soon as possible,” says Malavige, who, from May, is doubling the average number of samples sequenced each month, to nearly 200.
“We all have the question of why is it happening? Why is this wave now? And why is it so bad?” says Senjuti Saha, a molecular geneticist at the Child Health Research Foundation in Dhaka. New cases in Bangladesh reached more than 7,000 a day in mid-April. Numbers have fallen again since then, but Saha has noticed a big increase in sequences from the country shared on online genome databases and more labs are getting involved, which she puts down to concerns over the spread of variants.
Identifying worrying variants will help governments to make decisions about responses and restrictions, and whether more aggressive interventions are needed, says Dibesh Karmacharya, executive director of the Center for Molecular Dynamics Nepal, a non-profit organization in Kathmandu that sequences SARS-CoV-2 genomes.
Especially worrying is B.1.617, which was first identified in the Indian state of Maharashtra, and which the World Health Organization has said is probably highly transmissible. It is spreading fast through India and might have the ability to evade some immunity from previous infections or vaccination. “Sequencing has always been important, but it is critical now, because we are getting to a stage where more and more new variants are cropping up,” says Gavin Smith, a viral evolutionary biologist at Duke–NUS Medical School in Singapore. “We need sequence data to see what is going on.”
Worrying variants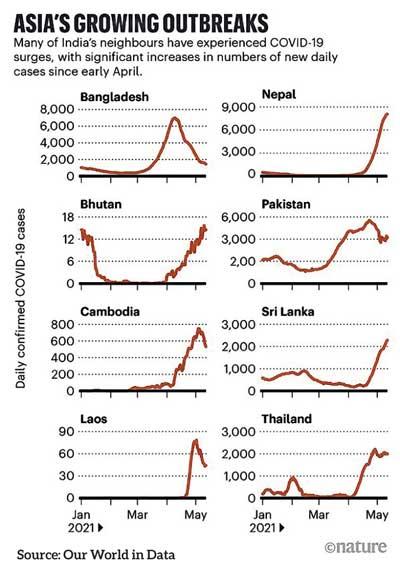
Sequenced samples in south and southeast Asia still represent only a small fraction of cases. But they are beginning to reveal a mixed picture of variants driving outbreaks in the region.
In Bangladesh, for example, the surge has coincided with the widespread detection of B.1.351, a variant first identified in South Africa, which has been linked to reduced effectiveness of the University of Oxford–AstraZeneca vaccine.
Saha first found the variant in her samples in February. In March, 28 of 36 genomes she sequenced were of B.1.351, as were more than 70% of 100 sequences shared by other groups in Bangladesh that month1. The variant has swept the country, and is especially worrying for Bangladesh, because most of the vaccines available there are Covishield, a version of the AstraZeneca vaccine produced in India.
The picture is different in Cambodia and Sri Lanka, where B.1.1.7 is predominant. Malavige wants to keep a close eye on its spread and evolution to see whether it develops a mutation known as E484K, which is present in some other variants and might allow it to evade immunity.
Cases in Cambodia have been rising steeply since early April. All 120 or so genomes sequenced there since then have been of B.1.1.7, says Erik Karlsson, a virologist at the Pasteur Institute of Cambodia in Phnom Penh, who runs the only lab in the country sequencing SARS-CoV-2.
The lab has greatly increased its sequencing capacity during the pandemic, and can now process more than two dozen genomes a week, providing near real-time insights. COVID-19 has “revolutionized the way we use sequencing”, says Karlsson, who has been sleeping at the institute to continue his work since a lockdown was imposed in Phnom Penh in mid-April.
The researchers in Bangladesh, Sri Lanka and Cambodia say they have not yet detected B.1.617 in their communities, but it could have crept in without being detected, owing to the minimal sequencing. “We are surrounded by India, and people are coming in and out all the time,” says Saha, and fewer than 1% of confirmed cases in Bangladesh have been sequenced.

Health workers administer SARS-CoV-2 tests at a railway station in Sri Lanka’s capital Colombo.Credit: Xinhua/eyevine
Frightening picture
To the northwest, B.1.617 has definitely made its way to Nepal, which shares an extensive, largely open border with India. Reported cases of COVID-19 have shot up since mid-April — hitting a new record of 9,317 cases on 11 May. Relative to population size, that’s more cases than in India.
In early May, Karmacharya’s team sequenced 12 samples from people recently infected in the Kathmandu valley — including himself. Eleven of the sequences were of B.1.617, and one was of B.1.1.7. The results are only a tiny snapshot of what is circulating, but are still “a little scary”, says Karmacharya, who had received one shot of a COVID-19 vaccine before getting infected and has now recovered.
But although researchers recognize that more genomic surveillance would be useful, their ambitions are limited by resources. “The main problem is a lack of funds,” says Malavige.
Karlsson points out that sequencing reagents are in high demand globally. With reduced airline activity, getting supplies to Cambodia “is a fight”.
Sequencing takes time to process, says Malavige. As a shortcut, her lab uses real-time polymerase chain reaction (PCR) kits that can quickly detect three widely circulating variants of concern: B.1.1.7, B.1.351 and P.1. But kits that detect B.1.617 have only just been developed and they cannot identify newly emerging variants.
“We are at a critical point where we need to keep sequencing to identify anything as soon as possible,” she says. And with vaccines still in very short supply in much of the region, many people will remain at risk, say researchers. Courtesy - www.nature.com
26 Nov 2024 26 Nov 2024
26 Nov 2024 26 Nov 2024
26 Nov 2024 26 Nov 2024
26 Nov 2024 26 Nov 2024
26 Nov 2024 26 Nov 2024